Spatial Technologies Unit
Spatial Technologies, including Spatial Transcriptomics and Proteomics, were deemed “Method of the Year 2020” by Nature Methods Journal and are currently revolutionizing biomedical research.
The Spatial Technologies Unit (STU) aims to become a center for innovation, efficient service provision, and a focal point for Spatial Technologies research and development in the Commonwealth of Massachusetts and across the United States. It hosts a comprehensive collection of leading spatial and single cell technologies from10x Genomics (Visium, Connect), NanoString (GeoMx), Akoya (Vectra Polaris, CODEX), Vizgen (MERSCOPE), and Bruker (Vutara VXL). Its unparalleled technological infrastructure, its location within the Harvard Medical School Ecosystem, and its translational role in a leading medical center provides exceptional opportunities for cutting-edge research and applications.
The STU provides a wide ranges of Spatial and Single Cell services to academia, startups, and industry, including:
- Access to super resolution microscopy on the Bruker Vutara VXL
- Full service single cell experiments (from tissue/cells to libraries and/or data) using the 10x Connect automated robotics platform.
- An extensive array of spatial transcriptomics and proteomic services, including:
- 10x Genomics Visium (for FFPE or frozen tissue sections)
- Akoya Biosciences Phenoptics (up to 9 antibodies on a whole slide (FFPE/frozen tissue), single cell resolution)
- Akoya CODEX (up to 60 antibodies on FFPE/tissue section, single cell resolution)
- NanoString GeoMx DSP (>18,000 genes or >1,000 proteins quantified on tissue sections/FFPE)
- Vizgen MERSCOPE (up to 500 genes quantified across a tissue section, subcellular resolution)
For further information on our services/pricing please contact us!
The Technology
10x Genomics Visium
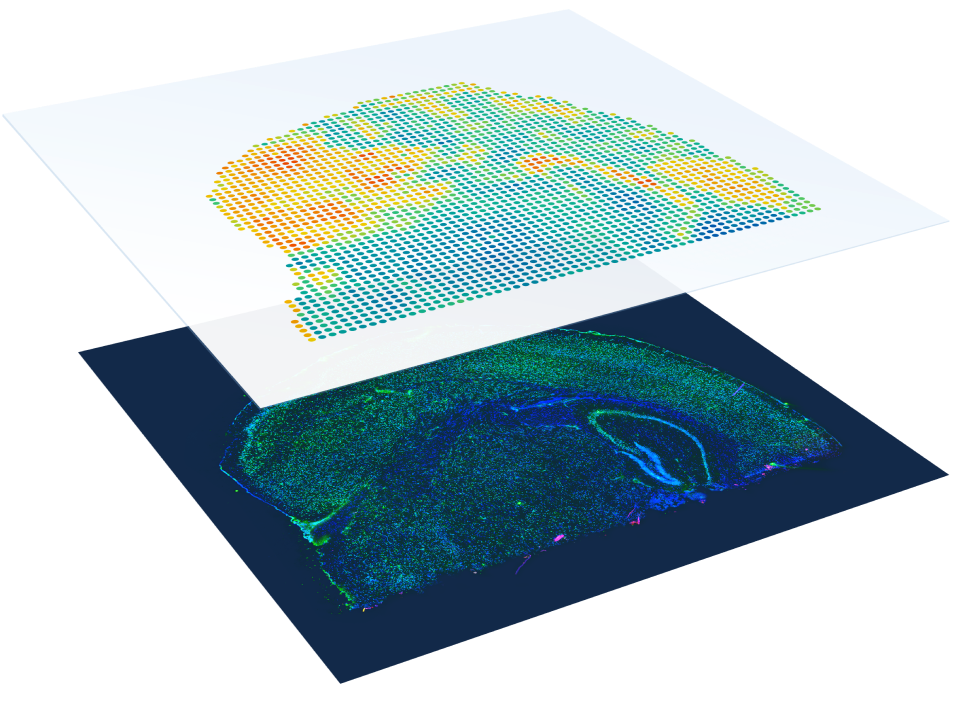
- Compatible with both FFPE and fresh frozen tissue samples
- No need to select regions of interest—analyze the whole transcriptome from an entire section.
- 1–10 cell resolution on average per spot depending on tissue type.
- Combine whole transcriptome spatial analysis with immunofluorescence protein detection.
The STU provides end-to-end service, from frozen tissue/paraffin blocks to libraries and/or data. We cover the tissue preparation, optimization, slide loading, visium experiment, library preparation, and sequencing.
For more information about the assay: https://www.10xgenomics.com/products/spatial-gene-expression
10x Genomics CONNECT

- Generate consistent results: Reduce single cell data variability, generating reproducible and consistent results.
- Characterize single cells by gene expression or immune receptor profiling.
- From sample to sequencing-ready libraries in one day
- Fast turnaround time and maximal consistency to all 10x genomics single cell assays: single nucleus sequencing, single cell sequencing, gene expression + TCR/BCR profiling (VDJ sequencing), and more!
See how the 10x Genomics Connect maximizes single cell robustness and reproducibility https://support.10xgenomics.com/single-cell-gene-expression/index/doc/technical-note-chromium-connect-consistent-automated-single-cell-gene-expression-library-generation
Please follow this link for more information on the instrument: https://www.10xgenomics.com/instruments/chromium-connect
Akoya Vectra Polaris

- State of the art multispectral imaging enables the identification and downstream quantification of multiple overlapping biomarkers (up to 8) without the interference of autofluorescence as the signals are unmixed from one another
- MOTiF™ technology generates unmixed whole slide scans at 40x magnification of up to 7 colors in about 20 minutes (15 x 15 mm region) for analysis of biology across the entire slide in a streamlined workflow and without selection bias.
- High speed, low-cost per slide, enabling screening of whole cohorts in a fraction of the cost and time
- Ability to work on protein level, capturing intracellular and surface protein markers
For more information: https://www.akoyabio.com/phenoptics/mantra-vectra-instruments/vectra-polaris/
Akoya CODEX
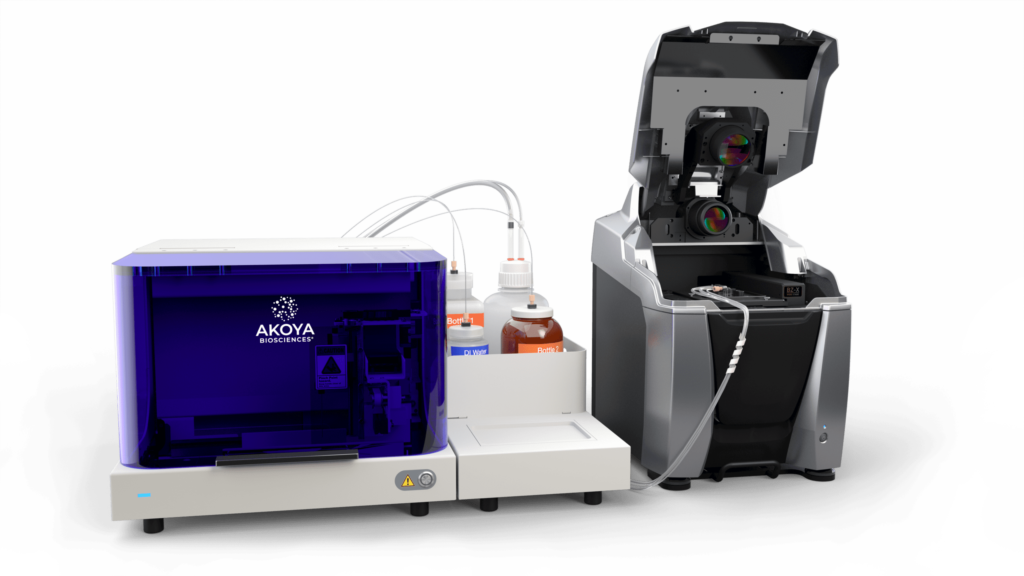
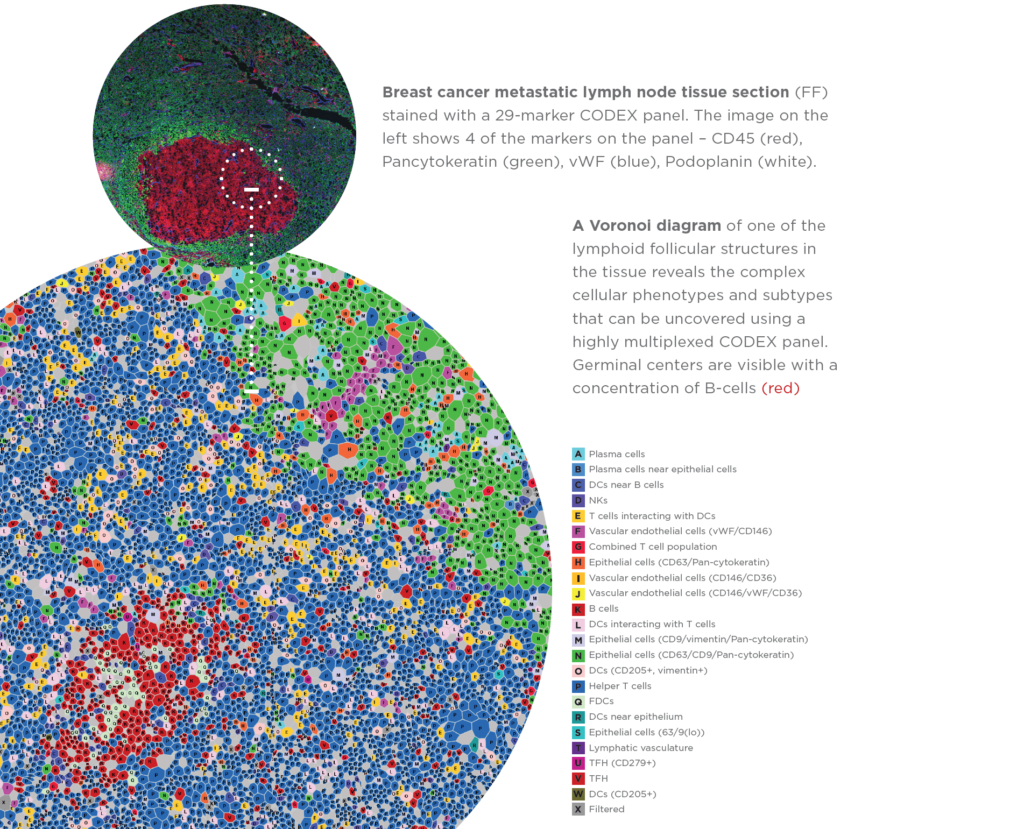
- Up to 60 antibodies imaged per slide at single cell resolution
- Create custom antibody panels for frozen tissue sections and FFPE
- Characterize in-depth cellular populations without requiring sequencing
- Ability to work on protein level, capturing intracellular and surface protein markers
Please follow this link for more information: https://www.akoyabio.com/codex/
NanoString GeoMx DSP

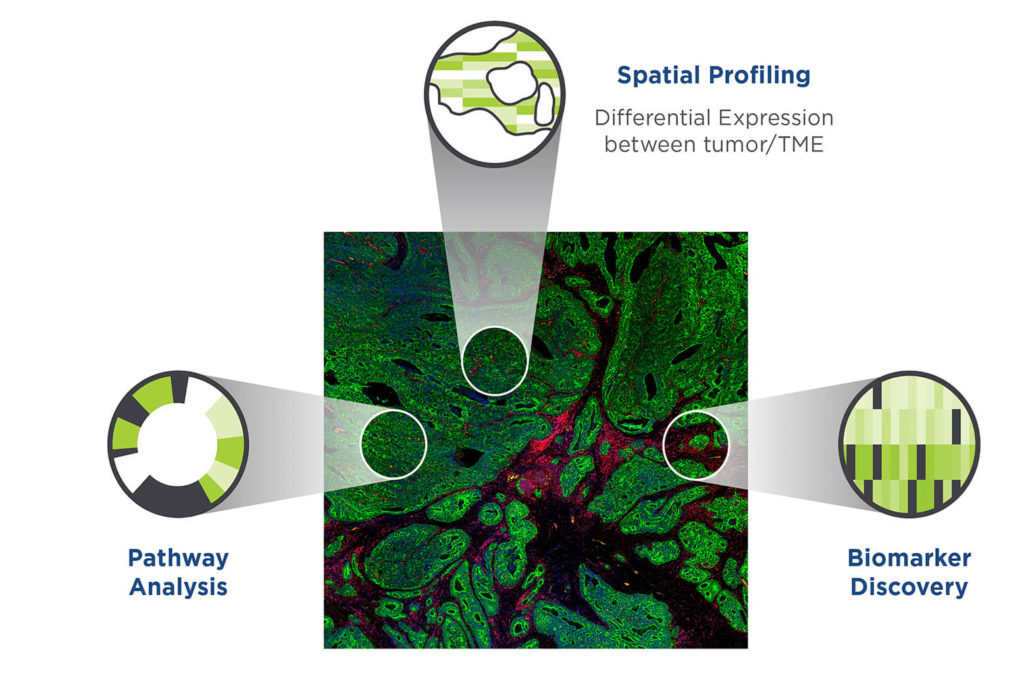
- Select up to 90 regions of interest (ROI) per tissue slide
- Ability to work seamlessly with FFPE or frozen tissue sections
- >18,000 genes quantified per ROI or >100 protein targets
- Ability to expand and include any target
For more information please visit: https://www.nanostring.com/products/geomx-digital-spatial-profiler/geomx-dsp-overview/
Vizgen MERSCOPE
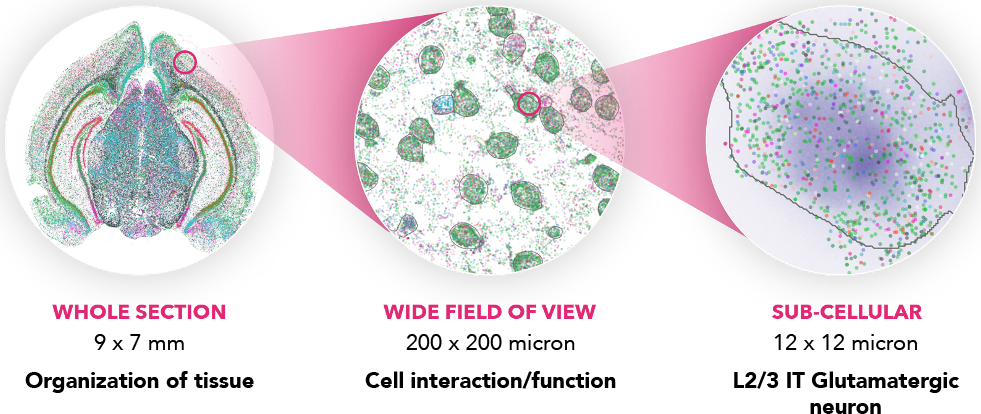
- Massively multiplexed, error-robust, single-cell in situ transcriptomic imaging
- Quantify and localize RNAs in any tissue sample easily, efficiently, and accurately
- Detect low expressing genes that can be missed using other technologies.
- Localize RNA transcripts at a subcellular level with ≤100 nm resolution.
- High multiplexing with custom gene panel design. Current chemistry up to 500 genes.
- Measure your specified genes on many sample or tissue types.
For more information please visit: https://vizgen.com/products/
Supporting Equipment
EPREDIA GrandMaster Tissue Microarrayer

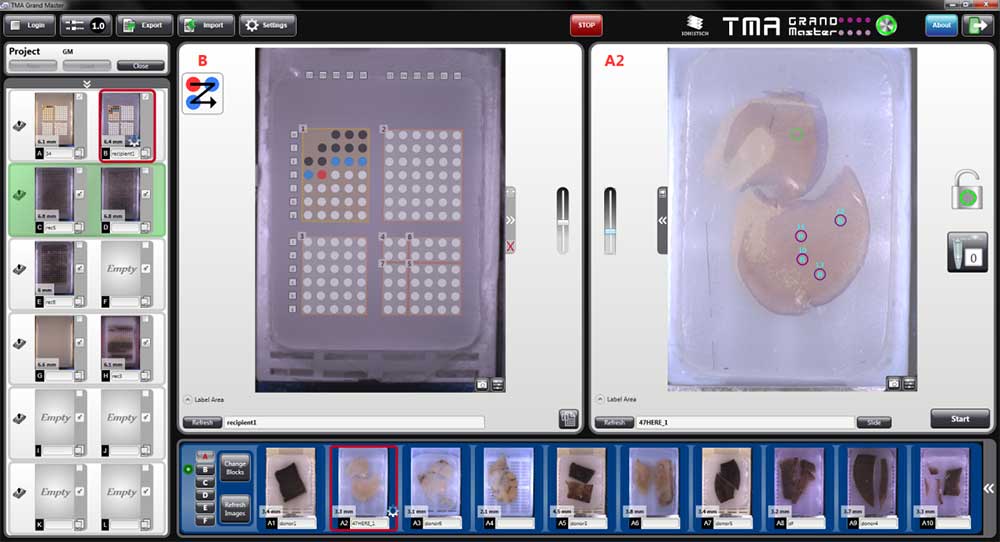
- The TMA Grand Master is the fastest, highest-capacity and most user-friendly tissue microarrayer on the market
- Allows the precise selection and extraction of specific regions of interest on the donor blocks and their insertion in recipient blocks
- It enables us to maximize the utility for the spatial technologies available in the STU and minimize costs by combining multiple regions, blocks, samples, and patients on the same slide. The TMA GrandMaster makes possible the screening of whole patient cohorts in just a handful of slides, minimizing costs and time.
- TMA blocks can be created by combining 0.6, 1, 1.5 and 2 mm diameter tissue cores. The instrument can also extract tissue samples to standard 0.2 ml PCR tubes. Then, the extracted tissue samples can be used later for various applications at the field of molecular pathology.
- 558 at 0.6mm, 286 at 1mm, 135 at 1.5mm, or 84 at 2mm tissue cores can be combined in a single block with unparalleled precision!
For more information please visit: https://www.3dhistech.com/research/tissue-microarrayers/tma-grand-master/
